Taverna is used by various projects and researchers for investigations into various diseases:
- Dementia (VPH-DARE@IT and VPH-SHARE)
- Analysis of the anthrax bacterium
- Study of resistance into trypanosomiasis (sleeping sickness)
- Identification of genes linked to Graves disease
- Characterisation of genes associated with Williams-Beuren syndrome
- Cancer research (caGrid)
Taverna has been used by Anil Wipat and others to automate a series of analyses on all the proteins produced by a bacterium to create, by a process of selection and elimination, a list of secreted proteins and their properties.
The secreted proteins explain why anthrax can grow in animal hosts but not in soil.
The paper e-Science Tools For The Genomic Scale Characterisation Of Bacterial Secreted Proteins by Craddock et al. gives an overview of this work.
Hide
One of the major goals of biology, and consequently bioinformatics, is to successfully bridge the gap between genotype and phenotype. Microarray and Quantitative Trait Loci data are increasingly used to aid in the discovery of candidate genes that might be responsible for phenotypic differences. In previous years, studies into genotype-phenotype correlations have been conducted manually. This has led to problems with regards to the identification of functional candidate genes, primarily due to the scale of data being investigated and the reliance on specific expertise, which may bias the investigations outcome.
With the development and connection of Web services into workflows, however, these large scale datasets can be processed systematically enabling detailed information to be gathered, published and subsequently re-investigated, enhancing the possibility of bridging the gap between genotype and phenotype using pathways.
So far this investigation has highlighted the issues facing the manual analysis of microarray and QTL data, and how automated approaches provide a systematic means to investigate genotype-phenotype correlations. We were able to illustrate how the large scale analysis of microarray gene expression and quantitative trait data, investigated at the level of biological pathways, enables links between genotype and phenotype to be successfully established.
An example of a workflow used to gather pathway information for candidate genes from a QTL region is shown below. These workflows have so far been applied to two different genotype-phenotype problems:
- The study of resistance to African Trypanosomiasis in mice, infected with Trypanosoma congolen
- The study of immunological effects and parasite expulsion in mice infected with Trichuris muris
Using this systematic, pathway driven approach, we were able to successfully identify a candidate gene and biological pathway believed to be strongly associated with resistance to African Trypanosomiasis.
The workflow for performing this analysis is available on myExperiment.
Publications
Articles and papers about the success of Taverna for trypanosomiasis (sleeping sickness) research.
Hide
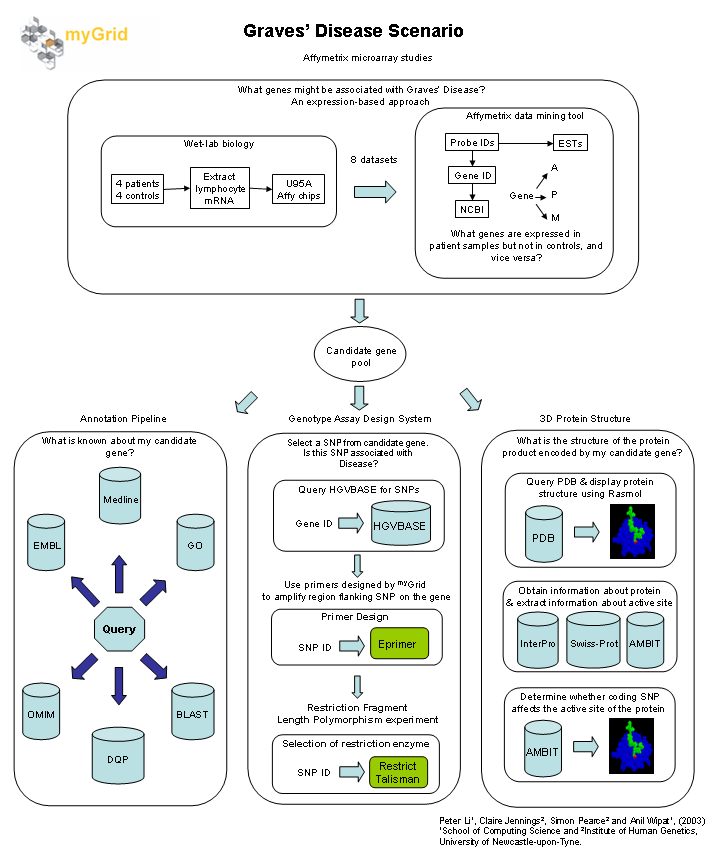
Graves Disease Scenario
The aim of this scenario is to identify and characterise genes which are located in regions on human chromosomes which show linkage to Graves disease (GD) (shown in figure below). GD is an autoimmune disease of the thyroid in which the immune system of an individual attacks cells in the thyroid gland resulting in hyperthyroidism. This is caused by the stimulation of the thyrotrophin receptor by thyroid-stimulating autoantibodies secreted by lymphocytes of the immune system.
Affymetrix microarray studies
The GD candidate genes were identified by microarray analysis. Affymetrix U95A arrays were probed with RNA extracted from CD4 positive lymphocytes from four GD patients and four healthy controls. The four GD microarray datasets were then compared to the four control datasets using the Affymetrix data mining tool to identify differentially expressed genes.
Annotation Pipeline
Over 50 genes were found to be differentially-expressed in CD4 positive lymphocytes from GD patients. In order to understand why these genes were expressed in lymphocytes from GD patients but not in healthy individuals, the GD biologist would like to use myGrid to query public databases such as EMBL, GO, HGVBASE and MEDLINE to view information about gene structure and function, chromosome location, the presence of single nucleotide polymorphisms (SNPs), expression control features and association with other genetic diseases. The experimental conditions and diseases in which the expression of the candidate genes are significantly altered also need to be identified from OMIM.
Genotype Assay Design System
SNPs are small (single base pair) changes genetic variations which are found in the genome amongst individuals. The differential expression of the candidate genes in GD individuals may be due or related to the presence of SNPs associated with GD. The GD biologist is interested in identifying and determining the frequency of those SNPs which are found in her GD patients.
Restriction fragment length polymorphism (RFLP) assays are developed to genotype SNPs in her candidate genes. A region flanking either side of the SNP is amplified using polymerase chain reaction (PCR). The amplified PCR product is digested with a suitable restriction enzyme (i.e. one that will cut at one SNP allele and not the other) and the products are run on agarose gels to view product size and determine the genotype.
The GD biologist would like to use myGrid to:
- Query databases to retrieve SNP information associated with candidate genes.
- Aid in the design of primers (bits of DNA which signify the start and end points of the section of the DNA sequence which she wants to amplify) for the PCR experiment.
- Select the restriction enzyme that is specific to a particular SNP for the RFLP experiment.
3D Protein Structure & effect of coding SNP on protein active site
Any SNPs occurring in the coding regions of a candidate gene may potentially give rise to a change in the amino acid sequence of the protein encoded by the gene. The GD biologist would like to use myGrid to:
- Query a protein structure database, e.g. PDB or MSD, to determine whether a structure of the protein encoded by her candidate gene is available. If so, view the protein structure to study how it relates to the function of the protein.
- Obtain information about the protein, e.g. its function and functional domains, by querying SWISS-PROT and InterPro. Use Sheffield’s AMBIT Web service to retrieve information about an active site whose characteristics may be altered due to the presence of a coding SNP which has affected a change in the amino acid sequence of the protein where the active site is encoded.
The workflow for the Graves Disease analysis is published on myExperiment.
Publications
Articles and papers about the success of Taverna for Graves Disease research.
Hide